One of the best outcomes of computational integration with biology is molecular docking. Molecular docking is a method that is used to determine the interaction between two molecules (protein-protein/ protein-ligand etc..).Molecular docking uses an algorithm which incorporates various other algorithms like search algorithm, which on the basis of small size and the moiety that fits within the protein’s cavity that predicts the ligand or in other words we can say docking determines the best orientation of ligand that forms complex with the protein along with best energy. The analysis is done on the basis of a scoring function which predicts the strength of the interaction (s) and converts the interacting energy into numerical form (docking score) which confirms the protein-ligand affinity.
There are different kinds of docking- Rigid (lock and key) and Flexible (induced fit). Docking is commonly used in drug designing which is also beneficial in lead remediation and bioremediation. Commonly and different softwares used in docking are- AutoDock, AutoDock Vina, PatchDock, Z-Dock,etc..
AutoDock by “The Scripp Research Institute” is a freely available software that includes an automated molecular docking procedure for the study of target-ligand interaction. This automated procedure consists of two programs one for calculating grids and other for docking the ligand with the calculate grid.there are two set of algorithm used –Lamarckian Genetic algorithm and an empirical free energy scoring function which can generate results for docking for upto 10 flexible bonds. Also, interaction can be seen pictorially for docked target-ligand complex as well.
AutoDock 4.2 is is the latest version which has now included limited side chain flexibility, atom types are expanded, unbound state, a desolvation model and force field. It has related software, that is, AutoDock Vina.
AutoDock Vina is software, that uses simple scoring function as compared to AutoDock and it can generate results for large system (20 flexible bonds.) as a result, average accuracy of binding is better than AutoDock 4.
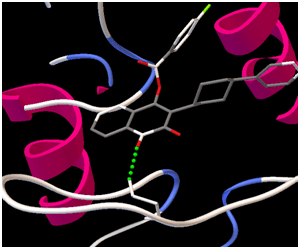
Image 1(a)
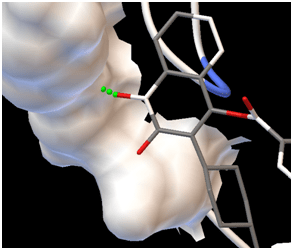
Image 2(b)
Image1 (a) & Image (b) : images showing interaction between target-ligand with AutoDock Tools.
With AutoDock Tools, bond flexibility in cyclic molecules cannot be managed in one step, which results to rigid ligand. Although with few methods these draw3bags can be overshadowed, like- docking can be done individually with identification of one or more low energy conformation or using the ring as a full entity and autodock converts it into acyclics form(after docking the cuclic form is restored))
Reference:
https://link.springer.com/protocol/10.1007%2F978-1-59745-177-2_19
